[ad_1]
Research
New AI system designs proteins that successfully bind to target molecules, with potential to advance drug design, disease understanding and more.
Every biological process in the body, from cell growth to immune response, depends on interactions between molecules called proteins. Like a key to a lock, one protein can bind to another and help regulate critical cellular processes. Protein structure prediction tools like AlphaFold have already given us tremendous insight into how proteins interact with each other to carry out their functions. However, these tools cannot create new proteins to directly manipulate these interactions.
However, scientists can create novel proteins that successfully bind to target molecules. These binders can help researchers accelerate progress across a wide range of research, including drug development, cell and tissue imaging, disease understanding and diagnosis – even plant resistance to pests. Although recent machine learning approaches to protein design have made great strides, the process is still laborious and requires extensive experimental testing.
Today we introduce AlphaProteo, our first AI system for developing novel, high-strength protein binders that serve as building blocks for biological and health research. This technology has the potential to accelerate our understanding of biological processes and support the discovery of new drugs, the development of biosensors, and more.
AlphaProteo can generate new protein binders for various target proteins, including VEGF-A, which is linked to cancer and complications from diabetes. This is the first time that an AI tool has been able to develop a successful protein binder for VEGF-A.
AlphaProteo also achieves higher experimental success rates and 3- to 300-fold better binding affinities than the best existing methods on seven target proteins we tested.
Learn the complicated ways proteins bind together
Protein binders that can bind tightly to a target protein are difficult to develop. Traditional methods are time-consuming and require multiple rounds of extensive laboratory work. After the binders are created, they go through further rounds of testing to optimize the binding affinity so that they bind tightly enough to be useful.
Based on large amounts of protein data from the Protein Data Bank (PDB) and more than 100 million predicted structures from AlphaFold, AlphaProteo has learned the countless ways molecules bind together. Given the structure of a target molecule and a set of preferred binding sites on that molecule, AlphaProteo generates a candidate protein that binds to the target at these sites.
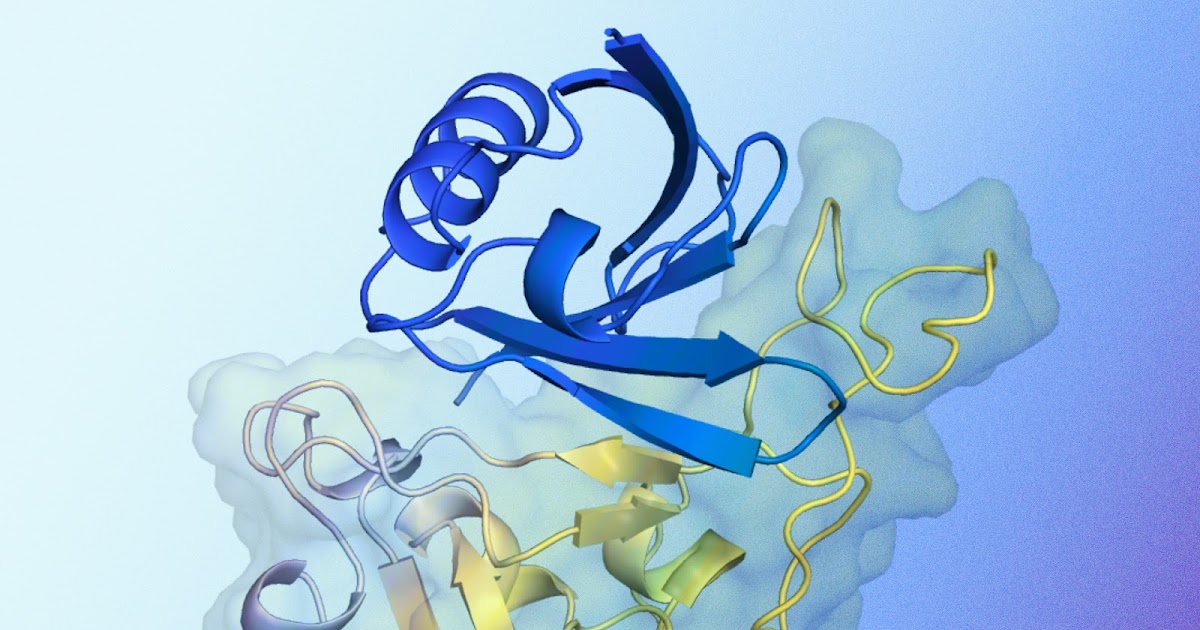
Illustration of a predicted protein binder structure interacting with a target protein. Shown in blue is a predicted protein binding structure generated by AlphaProteo that is designed to bind to a target protein. The target protein, in particular the SARS-CoV-2 spike receptor binding domain, is shown in yellow
Evidence of success at key protein binding targets
To test AlphaProteo, we developed binders for various target proteins, including two viral proteins involved in infections, BHRF1 and the spike protein receptor binding domain of SARS-CoV-2, SC2RBD, and five proteins that involved in cancer, inflammation and autoimmune diseases, IL-7Rɑ, PD-L1, TrkA, IL-17A and VEGF-A.
Our system has highly competitive retention success rates and best-in-class retention strengths. For seven targets, AlphaProteo generated candidate proteins in silico that bound strongly to the intended proteins in experimental tests.
A grid depicting predicted structures of seven target proteins for which AlphaProteo has successfully generated binders. Examples of binders tested in the wet laboratory are shown in blue, protein targets are shown in yellow, and intended binding regions are highlighted in dark yellow.
For one particular target, the viral protein BHRF1, 88% of our candidate molecules bound successfully when tested in the Google DeepMind Wet Lab. Based on the targets tested, AlphaProteo binders also bind on average ten times stronger than the best existing design methods.
For another target, TrkA, our binders are even stronger than the best previously developed binders for this target that have undergone multiple rounds of experimental optimization.
Bar graph showing the in vitro experimental success rates of the AlphaProteo output for each of the seven target proteins compared to other design methods. Higher success rates mean fewer designs need to be tested to find successful binders.
Bar graph showing the best affinity for AlphaProteo's designs without experimental optimization for each of the seven target proteins compared to other design methods. A lower affinity means that the binding protein binds more strongly to the target protein. Please note the logarithmic scale of the vertical axis.
Validation of our results
Over and beyond in silico To validate and test AlphaProteo in our wet lab, we commissioned the research groups of Peter Cherepanov, Katie Bentley and David LV Bauer from the Francis Crick Institute to validate our protein binders. In various experiments, they delved deeper into some of our stronger SC2RBD and VEGF-A binders. The research groups confirmed that the binding interactions of these binders were indeed similar to those predicted by AlphaProteo. In addition, the groups confirmed that the binders have a useful biological function. For example, some of our SC2RBD binders have been shown to block SARS-CoV-2 and some of its variants from infecting cells.
AlphaProteo's performance shows that it could dramatically reduce the time needed for initial experiments with protein binders for a wide range of applications. However, we know that our AI system has limitations as it was unable to develop successful binders against an 8th target, TNFɑ, a protein associated with autoimmune diseases such as rheumatoid arthritis. We chose TNFɑ to seriously challenge AlphaProteo because computational analysis showed that it would be extremely difficult to design binders against it. We will continue to improve and expand AlphaProteo's capabilities to ultimately address such challenging goals.
Achieving strong binding is usually only the first step in developing proteins that could be useful for practical applications, and many more biotechnological obstacles must be overcome in the research and development process.
Towards responsible protein design development
Protein design is a rapidly evolving technology that holds great potential for advancing science in everything from understanding the factors that cause disease to accelerating the development of diagnostic tests for viral outbreaks to supporting more sustainable manufacturing processes and even removing pollutants from the environment .
To address potential risks in biosecurity, building on our long-standing approach to responsibility and safety, we are working with leading external experts to inform our phased approach to sharing this work and to integrate community efforts to develop best practices, including the , to flow into the new AI Bio Forum from NTI (Nuclear Threat Initiative).
In the future, we will work with the scientific community to use AlphaProteo on important biological problems and to understand its limitations. We at Isomorphic Labs have also been exploring its applications in drug design and are excited about the future.
At the same time, we continue to improve the success rate and affinity of AlphaProteo's algorithms, expand the range of design problems it can solve, and collaborate with researchers in machine learning, structural biology, biochemistry and other disciplines to provide a responsible and responsible solution develop more comprehensive protein design offerings for the community.
If you are a biologist whose research could benefit from target-specific protein binding and would like to express interest in becoming a trusted tester for AlphaProteo, please contact alphaproteo@google.com.
We will process the received messages according to our Privacy Policy.
Acknowledgments
This research was co-developed by our Protein Design team and our Wet Lab team.
We would like to thank our collaborators Peter Cherepanov, David Bauer, Katie Bentley and their groups at the Francis Crick Institute for their invaluable experimental insights and results, the AlphaFold team whose previous work and algorithms provided training inputs and evaluation insights, and the many other teams at Google DeepMind, who contributed to this program.
[ad_2]
Source link